MOLEonline 2.0 introduction page
MOLEonline 2.0 web interface provides a direct access to MOLE 2.0 functionality and enables on-line and easy-to-use interactive channel analysis.
MOLE 2.0 is an universal toolkit for rapid and fully automated location and characterization of channels, tunnels and pores in (bio)macromolecular structures, e.g., proteins, RNA, DNA and biomacromolecular assemblies.
MOLEonline 2.0 features
- Quickest channel calculation on the market
- Channel profile - geometry
- List of residues lining channels (distinguishing sidechain/mainchain contact with the channel)
- Physico-chemical properties (charge, polarity, hydropathy, hydrophobicity, mutability, etc.) of channel
- Automatic active site identification for enzymes using CSA
- Interactive visualization of structure and channels with Jmol
- Automatic detection of starting points in cavities
- Interactive selection of starting points
- Visualization of starting points
- Cavity and molecular surface visualization
- Experimental feature - pore identification
- Download results - as a report, PDB files and script for PyMol®
- Free of charge
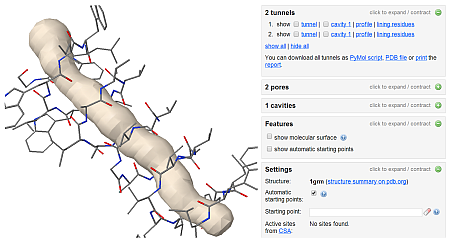
Example of working environment with gramicidin (1GRM).
Quick start
News
beta.mole.upol.cz
01. 04. 2017MOLEonline with integrated Litemol vizualization overcomes current problems of tunnel visualization in Jmol and brings some new functionality. Try it at http://beta.mole.upol.cz/. We are developing completely new version. It will be released by the end of 2017.
ELIXIR CZ support
01. 01. 2016MOLEonline is now supported by ELIXIR CZ research infrastructure project (MEYS Grant No: LM2015047).
Java problems
21. 11. 2014If you encounter "security" problems with new Java, go to the Java
control panel and lower the security level to medium.